 PDF(6056 KB)
PDF(6056 KB)


 PDF(6056 KB)
PDF(6056 KB)
 PDF(6056 KB)
PDF(6056 KB)
基于GANBPSO-SVM的高光谱影像特征选择方法
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}A Feature Selection Strategy for Hyperspectral Images Classification Based on GANBPSO-SVM
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}为了在保持对目标检测和分类分析所需信息的同时,降低高光谱影像的维度,提出了一种混合优化策略的特征选择方法。该方法将遗传算法和二进制粒子群优化算法融合成一种新的混合优化策略(GANBPSO),自动选择最优波段组合,同时优化分类器支持向量机(RBF-SVM)的参数,以提高分类器的分类性能。为了说明所提出方法的有效性,采用了在高光谱分类中广泛使用的Indian Pines(AVIRIS 92AV3C)数据集进行测试。结果表明所提出方法(GANBPSO-SVM)能够自动选择包含最多信息的特征子集以保证分类精度,而不需要预先设置所需要的特征子集数量,本方法与传统方法相比具有更好的分类效果。
Rich spectral information from hyperspectral images can aid in the classification and recognition of the ground objects. Currently, hyperspectral images classification has already been applied successfully in various fields. However, the high dimensions of hyperspectral images cause redundancy in information and bring some troubles while classifying precisely ground truth. Hence, this paper proposes a hybrid feature selection strategy based on the Genetic Algorithm and the Novel Binary Particle Swarm Optimization (GANBPSO) to reduce the dimensionality of hyperspectral data while preserving the desired information for target detection and classification analysis. The proposed feature selection approach automatically chooses the most informative features combination. The parameters used in support vector machine (SVM) simultaneously are optimized, aiming at improving the performance of SVM. To show the validity of the proposal, Indian Pines(AVIRIS 92AV3C) data set which is widely used to test the performance of feature selection techniques is chosen to feed the proposed method. The obtained results clearly confirm that the new approach is able to automatically select the most informative features in terms of classification accuracy without requiring the number of desired features to be set a priori by users. Experimental results show that the proposed method can achieve higher classification accuracy than traditional methods.
高光谱影像 / 特征选择 / 粒子群优化 / 支持向量机 {{custom_keyword}} /
hyperspectral images / feature selection / Particle Swarm Optimization / Support Vector Machine {{custom_keyword}} /
表1 训练集和测试集Table 1 Number of training and test samples |
| 类别号 | 类别名称 | 训练集 | 测试集 |
|---|---|---|---|
| 1 | 苜蓿 | 5 | 41 |
| 2 | 免耕玉米 | 143 | 1285 |
| 3 | 少耕玉米 | 83 | 747 |
| 4 | 玉米地 | 24 | 213 |
| 5 | 牧场 | 49 | 434 |
| 6 | 树 | 73 | 657 |
| 7 | 收割牧草 | 3 | 25 |
| 8 | 干草匀堆料 | 48 | 430 |
| 9 | 燕麦 | 2 | 18 |
| 10 | 免耕大豆 | 98 | 874 |
| 11 | 少耕大豆 | 246 | 2209 |
| 12 | 净耕大豆 | 60 | 533 |
| 13 | 小麦 | 21 | 184 |
| 14 | 木材 | 127 | 1138 |
| 15 | 建筑-草地-树木-机器 | 39 | 347 |
| 16 | 石钢塔 | 10 | 83 |
| 共计 | 1031 | 9218 |
表2 KC统计值与分类效果对应关系Table 2 Classification quality associated to the Kappa statistics value |
| KC | 分类效果 |
|---|---|
| <0 | 较差 |
| 0~0.2 | 差 |
| 0.2~0.4 | 正常 |
| 0.4~0.6 | 好 |
| 0.6~0.8 | 较好 |
| 0.8~1.0 | 非常好 |
表3 不同分类器200个波段下的分类精度Table 3 Classification accuracy by different classifiers in the original hyperspectral image |
| 分类方法 | 参数范围 | OA(%) | KC |
|---|---|---|---|
| ELM | N∈[5,50] | 71.09 | 0.6873 |
| KNN | K∈[1,15] | 73.92 | 0.6920 |
| SVM | C=100,γ=2 | 79.04 | 0.7611 |
| C=120,γ=2 | 78.74 | 0.7588 | |
| C=200,γ=6 | 81.80 | 0.7930 |
表4 特征选择和SVM参数优化前后的分类精度Table 4 Classification accuracy of SVM parameters optimization |
| 分类方法 | 波段数 | 参数 | OA(%) | KC |
|---|---|---|---|---|
| SVM | 200 | C=200,γ=6 | 81.80 | 0.7930 |
| GANBPSO-SVM | 44 | C=241.6169,γ=8.1961 | 85.56 | 0.8323 |
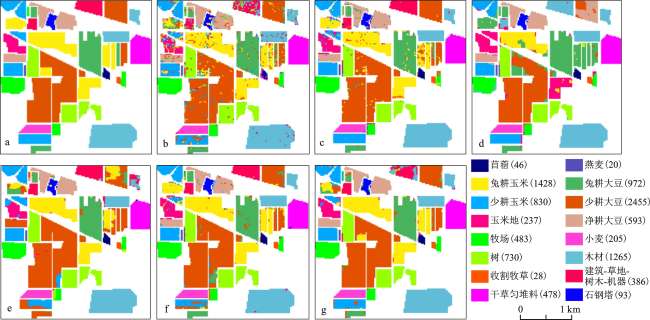
图3 分类结果对比 |
表5 基于SVM的不同特征选择方法的分类精度Table 5 classification accuracy by different feature selection algorithms based on SVM |
表6 基于GANBPSO-SVM的不同比例训练样本的分类结果Table 6 Classification accuracy based on GANBPSO-SVM under different proportions of the training samples |
| 类别 | 训练样本比例(%) | |||
|---|---|---|---|---|
| 10 | 20 | 50 | 80 | |
| OA(%) | 85.56 | 87.99 | 91.04 | 94.61 |
| KC | 0.8323 | 0.8623 | 0.8946 | 0.9384 |
| [1] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [2] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [3] |
[
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [4] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [5] |
[
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [6] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [7] |
[
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [8] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [9] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [10] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [11] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [12] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [13] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [14] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [15] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [16] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [17] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [18] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [19] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [20] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [21] |
Landgrebe D A.ftp:MultiSpec/ ThyFiles Indiana’s Pines Dataset[EB/OL].2009-08-10. //ftp. ecn. purdue. edu/ biehl/ PCzip (ground truth).
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| [22] |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
The authors have declared that no competing interests exist.
 PDF(6056 KB)
PDF(6056 KB)
 图1 粒子编码
图1 粒子编码 表1 训练集和测试集
表1 训练集和测试集 图2 高光谱数据集(a)和相应的参考数据(b)图例括号中数字为像素点个数。
图2 高光谱数据集(a)和相应的参考数据(b)图例括号中数字为像素点个数。 表2 KC统计值与分类效果对应关系
表2 KC统计值与分类效果对应关系 表3 不同分类器200个波段下的分类精度
表3 不同分类器200个波段下的分类精度 表4 特征选择和SVM参数优化前后的分类精度
表4 特征选择和SVM参数优化前后的分类精度 图3 分类结果对比图例括号中数字为像素点个数。a.参考数据; b. PSO- SVM分类结果(10%训练样本); c. HGAPSO- SVM分类结果(10%训练样本); d. GANBPSO-SVM(10%训练样本)分类结果; e. GANBPSO-SVM(20%训练样本)分类结果; f. GANBPSO-SVM(50%训练样本)分类结果; g.GANBPSO-SVM(80%训练样本)分类结果
图3 分类结果对比图例括号中数字为像素点个数。a.参考数据; b. PSO- SVM分类结果(10%训练样本); c. HGAPSO- SVM分类结果(10%训练样本); d. GANBPSO-SVM(10%训练样本)分类结果; e. GANBPSO-SVM(20%训练样本)分类结果; f. GANBPSO-SVM(50%训练样本)分类结果; g.GANBPSO-SVM(80%训练样本)分类结果 表5 基于SVM的不同特征选择方法的分类精度
表5 基于SVM的不同特征选择方法的分类精度 表6 基于GANBPSO-SVM的不同比例训练样本的分类结果
表6 基于GANBPSO-SVM的不同比例训练样本的分类结果/
| 〈 |
|
〉 |